AbstractA total of 16 Taenia multiceps isolates collected from naturally infected sheep or goats in Gansu Province, China were characterized by sequences of mitochondrial cytochrome c oxidase subunit 1 (cox1) gene. The complete cox1 gene was amplified for individual T. multiceps isolates by PCR, ligated to pMD18T vector, and sequenced. Sequence analysis indicated that out of 16 T. multiceps isolates 10 unique cox1 gene sequences of 1,623 bp were obtained with sequence variation of 0.12-0.68%. The results showed that the cox1 gene sequences were highly conserved among the examined T. multiceps isolates. However, they were quite different from those of the other Taenia species. Phylogenetic analysis based on complete cox1 gene sequences revealed that T. multiceps isolates were composed of 3 genotypes and distinguished from the other Taenia species.
INTRODUCTIONCoenurosis, caused by the larval form of Taenia multiceps, is a parasitic disease of various livestock species, especially ruminants. Coenurosis is common in sheep and goats worldwide and also has public health concerns because of its infection in humans [1,2]. T. multiceps larval cysts are usually found in nervous system, including brain and spinal cord of many herbivores. The presence of cysts typically leads to neurological symptoms that cause the infected animals to die after some weeks from starvation. However, animals in most cases remain normal without clinical symptoms and the disease is diagnosed only after the death of the animals. T. multiceps is a taeniid cestode whose adult stage lives in the small intestine of definitive hosts such as dogs and other canids [3]. T. multiceps larvae infection was reported in countries where sheep and goats herding remain important components of local economies, including some areas of Europe, Africa, Americas, and Asia. In China, T. multiceps infection was reported in dogs in several provinces with infection rate between 2.3% and 14.6% [4,5]. Although coenurosis in animals was reported in more than 20 provinces of China, molecular identification of T. multiceps isolates was rarely recorded. Moreover, the molecular identification of T. multiceps isolates is of great importance for etiological, epidemiological, and phylogenetic studies.
Mitochondrial (mt) DNA is known to have a faster evolutionary rate than nuclear DNA and is generally inherited maternally in almost all metazoans [6,7]. The mt genome is considered to be clonal and rarely or never undergoes recombination. Sequences generated from the mt genome provide excellent molecular markers for defining population groups, for tracing the genetic history of an individual or a particular group of related individuals, and for reconstructing deep-branch taxonomic phylogenies. With respect to the molecular identification of cestodes, early studies using partial cytochrome c oxidase subunit 1 gene (cox1) and NADH dehydrogenase-1 gene helped to establish the population and genetic relationship of T. multiceps and other members of the Taeniidae [8-10]. However, to our knowledge, there have been rare reports on molecular characterization of T. multiceps isolates in China. For this reason, we described in this paper an investigation of genetic variability among 16 T. multiceps isolates from Gansu Province, China by using sequences of cox1 gene as genetic marker.
MATERIALS AND METHODSCollection of T. multiceps isolatesBetween May 2008 and June 2010, a total of 16 isolates of T. multiceps larval stage were obtained from naturally infected sheep or goats in Gansu Province which is situated in the northwestern part of China between the northern latitudes of 32° to 40° and eastern longitudes of 94° to 108°. Sample code, geographical origin, and host are shown in detail in Table 1. The protoscolices were removed from individual larval cysts, washed 3 times with saline and stored at -70℃ before extraction of genomic DNA.
Genomic DNA extraction and amplification by PCRGenomic DNA was extracted from protoscolices using Genomic Tissue DNA Kit (Omega Biotek, Norcross, Georgia, USA) according to the manufacturer's recommendations. Genomic DNA concentrations were determined spectrophotometrically by GeneQuant 100 and the DNA samples were stored at -20℃ until used. The primers to amplify the complete cox1 gene were designed according to the mitochondrial genome sequence of T. multiceps [11] as follows: PF 5'-GACTTTGTTTGAGTCATCTTATGTAA-3' and PR 5'-GATTTACAAAATCTACATTTTAAA-3'. PCR (50 µl) was performed in 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM of each dNTP, 20 pmol of each primer and 2 U Taq DNA polymerase (Takara, Tokyo, Japan) under the following cycling conditions: after an initial denaturation at 94℃ for 5 min, then 94℃ for 1 min (denaturation), 55℃ for 1 min (annealing), 72℃ for 1min (extension) for 35 cycles, followed by a final extension at 72℃ for 10 min. Samples without genomic DNA were included in each amplification run as negative controls. PCR products were separated on a 1% agarose gel and detected by ethidium bromide staining.
DNA sequencing and analysisAll of the amplicons were ligated into pMD-18 T vector (Takara) and transformed to Escherichia coli DH5α competent cells. The positive colonies were sequenced with ABI PRISM 3730 Genetic Analyzer, and sequences were analyzed with DNAtools software. Further comparison with taeniid cox1 gene sequences available in GenBank was made using the BLAST and Clustal W software. Phylogenetic analyses were performed using the neighbor joining (NJ), maximum parsimony (MP), and minimum evolution (ME) methods, and the Mega 5.05 program. The bootstrap value was set as 1,000 replications with a cutoff value of 50%.
RESULTSPCR amplification of cox1 geneIn the present study, 1 to 3 larval cysts were observed in the brain of naturally infected sheep, while 7 cysts were found in the brain of the infected goat. The characteristics and morphology of T. multiceps larvae were seen in all of the cysts containing clear fluid with numerous fertile protoscolices (Fig. 1). Using 16 isolates of T. multiceps protoscolices DNA as templates, a band of approximately 1.7 kb fragment was generated after PCR amplification of cox1 genes. Each of the PCR products was ligated to pMD-18 T vector, and 3 colonies for each sample were sequenced on both direction and the consensus sequences were obtained.
Sequence analysis of cox1 geneAfter removal of the primer sequences, the cox1 gene from each isolate was 1,623 bp in length encoding 540 amino acids. Sequencing results indicated that out of 16 T. multiceps isolates 10 unique cox1 gene sequences were obtained with sequence variation of 0.12-0.68% and a total of 19 nucleotide variation sites. Some of the isolates had identical cox1 gene sequences, such as isolates JT081204 and JT090318, isolates JT090104 and PL090603, isolates JT09115-3 and JT090603, isolates YJ100610 and ZJCH100610, and isolates JT100124-1, JT100124-2 and JT100202. The G+C contents of the cox1 gene sequences of T. multiceps isolates were 29.9-30.4%. The percent identities among T. multiceps isolates from Gansu Province were more than 99% and showed a lesser degree of variability. Jingtai isolates JT081204 and JT090318 were identical with the corresponding sequence available in GenBank (GQ228818). When the 10 unique cox1 gene sequences were translated into amino acids sequence, 7 unique protein sequences were obtained with sequence variation of 0.19-0.56% and a total of 6 amino acid variation sites. Amino acid sequences of isolates JT080526, JT090104, JT090115-2, and YJ100610 were identical to the corresponding sequence available in GenBank (ACK55903) and that of isolate JT081204 was identical to T. multiceps
cox1 available in GenBank (ACS37254).
When Blast analysis with cox1 gene sequences was performed, more than 20 T. multiceps partial cox1 gene sequences from 346 to 445 bp were also available in GenBank. Partial cox1 gene sequences from the 10 unique sequences in this study could be further grouped into 4 unique sequences with variation rate of 0.25-0.75%. The first group included JT080526, JT090104, and JT100124-1, and was identical to the Turkish T. multiceps isolate tmtr01 (EF393620). The second group consisted of JT081204, JT090115-3, and JT090331, and was identical to the Italian T. multiceps isolate Tm2 (DQ309768). The third group included JT081008, JT090115-2, and YJ100610, and the last group was JT090115-1. Comparison of all the partial cox1 gene sequences revealed 10 groups with variation rate of 0.25-4.55%.
Phylogenetic analysis of taeniid cox1 geneThe complete cox1 gene sequences from the other Taenia mt genomes available in GenBank were selected and aligned. Accordingly, a phylogenetic tree was constructed with cox1 gene sequences from this study and previously published mt sequences of related Taenia species, including T. multiceps (GQ228818 and FJ495086), T. saginata (AY684274), T. asiatica (AF445798), T. solium (AB086256), T. hydatigena (GQ228819 and FJ518620), T. crassiceps (AF216699), T. pisiformis (GU569096 and NC013844), and T. taeniaeformis (JQ663994 and FJ597547), with E. granulosus (AF297617) as the outgroup. The cox1 gene sequences were 1,623 bp in length for all T. multiceps isolates, but it was 1,620 bp for T. saginata, T. asiatica, T. solium, T. hydatigena, and T. pisiformis. However, it was 1,614 bp and 1,635 bp for T. crassiceps and T. taeniaeformis, respectively.
Phylogenetic reconstruction of cox1 gene from Taenia species using the NJ, MP, and ME analysis showed similar topology; 3 genotypes were revealed in the examined T. multiceps isolates (Fig. 2). The first genotype consisted of JT090115-3, JT090115-1, T. multiceps GQ228818, JT081204, and JT090331. The second genotype included JT081008, JT090115-2, and YJ100610. The third genotype comprised of JT090104, JT101124-1, JT080526, and T. multiceps FJ495086. The cox1 gene sequences of T. multiceps were more similar to those of T. saginata and T. asiatica, suggesting that the cox1 gene could be a good genetic marker for molecular identification of Taenia species.
DISCUSSIONCestodes of the family Taeniidae (taeniids) occur as adult tapeworms in the small intestine of carnivorous definitive hosts, and are transmitted to specific intermediate mammalian hosts where they develop as fluid-filled metacestodes in tissues. A number of Taenia species including T. multiceps are of medical and/or economic significance [12]. For the molecular characterization and phylogeny study of taeniid cestodes, many genetic markers have been used, including 28S rDNA, cox1, nad1, nad4, ITS rDNA, and nuclear protein coding genes such as rpbz, pepck, and pold [8-10,13-18]. The cox1 gene is one of the most used markers for Taenia identification. Varcasia et al. [19] analyzed genetic variation within 40 T. multiceps isolates obtained from various locations of Sardinia. Partial sequences of nad1 and cox1 showed differences ranging from 1.27 to 2.54% and from 0.22 to 0.67%, respectively, and a conclusion was made that Sardinian sheep samples had at least 3 specific genetic variants [19]. However, comparison between the partial cox1 sequences of the T. multiceps isolates from infected cattle from Erzurum Province, Turkey and other T. multiceps isolates available in GenBank showed nucleotide differences ranging from 0.2 to 2.6% [20].
Although coenurosis in sheep and goats were reported in China, the molecular identification of T. multiceps was rarely conducted. Recently, the complete mt genome of T. multiceps from Chinese isolates was reported and provided useful markers for studying the systematics, population genetics, and molecular epidemiology of Taenia [13,21]. Partial cox1 genese of 3 larval isolates from goats in Hunan Province were sequenced and showed 97.9% identity to the T. multiceps
cox1 gene (EF393620), indicating that these parasites in goats belong to the larvae of T. multiceps [22]. The partial cox1 gene sequences in this study could be divided into 3 types with variation rates of 0.25 to 0.75%, which is similar to the previous report [19]. The phylogenetic tree based on cox1 gene sequences showed that all T. multiceps isolates constitute a branch consisting of 3 genotypes. T. multiceps, T. saginata, and T. asiatica showed the closest relationships, followed by T. solium, T. pisiformis, T. hydatigena, T. crassiceps, and the other Taenia species. Therefore, the cox1 gene sequence can be used as a molecular marker for the interspecific identification of Taenia species.
When partial sequences of the cox1 gene between T. asiatica and T. saginata were compared, the sequence differences varied from 2% in the conserved region (nucleotide position of 742-1,107) to 7% in the variable region (positions 25-340) [23]. It was suggested that partial cox1 sequences might give a bias to analysis, but the complete sequence data should provide a more reliable result. In this study, T. multiceps metacestode (larval form) from naturally infected sheep or goat in 4 geographical origins in Gansu Province were genetically characterized for the first time with complete cox1 gene sequences. We have detected some polymorphisms within the common 'sheep' isolates. It is evident that intraspecific variation existed in T. multiceps, whether such minor genetic variation has biological or epidemiologic significance warrants further study. In addition, for studying the population genetics of T. multiceps from sheep and goats in China, more isolates should be collected and analyzed. Further investigations will require more complete cox1 gene sequences from T. multiceps isolates from different endemic regions in China and will determine whether there are morphological, developmental, or pathological differences between the genotypes.
ACKNOWLEDGMENTSThis work was supported by National Key Project of Scientific and Technical Supporting Program (No. 2007BAD40B04) and partially supported by National Beef Cattle and Yak Industrial Technology System, NBCITS, MOA (No. CARS-38); National Nonprofit Institute Research Grant (No. 1610322012026); Gansu Province Agricultural Biotechnology Research and Application Development Project (No. GNSW-2010-01); Gansu Province Scientific and Technical Supporting Program (No. 1104NKCA082).
REFERENCES1. Sharma DK, Chauhan PPS. Coenurosis status in Afro-Asian region: A review. Small Rumin Res 2006;64:197-202.
2. Collomb J, Machouart M, Biava MF, Brizion M, Montagne K, Plénat F, Fortier B. Contribution of NADH dehydrogenase subunit I and cytochrome C oxidase subunit I sequences toward identifying a case of human coenuriasis in France. J Parasitol 2007;93:934-937. PMID: 17918379.
3. Scala A, Varcasia A. Updates on morphobiology, epidemiology and molecular characterization of coenurosis in sheep. Parassitologia 2006;48:61-63. PMID: 16881398.
4. Wang CR, Qiu JH, Zhao JP, Xu LM, Yu WC, Zhu XQ. Prevalence of helminthes in adult dogs in Heilongjiang Province, the People's Republic of China. Parasitol Res 2006;99:627-630. PMID: 16715234.
5. Dai RS, Li ZY, Li F, Liu DX, Liu W, Liu GH, He SW, Tan MY, Lin RQ, Liu Y, Zhu XQ. Severe infection of adult dogs with helminths in Hunan Province, China poses significant public health concerns. Vet Parasitol 2009;160:348-350. PMID: 19091472.
6. Ballard JWO, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Annu Rev Ecol Evol Syst 2005;36:621-642.
7. Le TH, Blair D, McManus DP. Mitochondrial genomes of parasitic flatworms. Trends Parasitol 2002;18:206-213. PMID: 11983601.
8. Gasser RB, Zhu X, McManus DP. NADH dehydrogenase subunit 1 and cytochrome c oxidase subunit I sequences compared for members of the genus Taenia (Cestoda). Int J Parasitol 1999;29:1965-1970. PMID: 10961852.
9. Zhang L, Hu M, Jones A, Allsopp BA, Beveridge I, Schindler AR, Gasser RB. Characterization of Taenia madoquae and Taenia regis from carnivores in Kenya using genetic markers in nuclear and mitochondrial DNA, and their relationships with other selected taeniids. Mol Cell Probes 2007;21:379-385. PMID: 17600673.
10. Lavikainen A, Haukisalmi V, Lehtinen MJ, Henttonen H, Oksanen A, Meri S. A phylogeny of members of the family Taeniidae based on the mitochondrial cox1 and nad1 gene data. Parasitology 2008;135:1457-1467. PMID: 18937885.
11. Jia WZ, Yan HB, Guo AJ, Zhu XQ, Wang YC, Shi WG, Chen HT, Zhan F, Zhang SH, Fu BQ, Littlewood DT, Cai XP. Complete mitochondrial genomes of Taenia multiceps, T. hydatigena and T. pisiformis: additional molecular markers for a tapeworm genus of human and animal health significance. BMC Genomics 2010;11:447. PMID: 20649981.
12. Hoberg EP. Taenia tapeworms: their biology, evolution and socioeconomic significance. Microbes Infect 2002;4:859-866. PMID: 12270733.
13. Bowles J, McManus DP. Genetic characterization of the Asian Taenia, a newly described taeniid cestode of humans. Am J Trop Med Hyg 1994;50:33-44. PMID: 7905720.
14. Gasser RB, Chilton NB. Characterisation of taeniid cestode species by PCR-RFLP of ITS2 ribosomal DNA. Acta Trop 1995;59:31-40. PMID: 7785524.
15. de Queiroz A, Alkire NL. The phylogenetic placement of Taenia cestodes that parasitize humans. J Parasitol 1998;84:379-383. PMID: 9576516.
16. Eom KS, Chai JY, Yong TS, Min DY, Rim HJ, Kihamia C, Jeon HK. Morphologic and genetic identification of Taenia tapeworms in Tanzania and DNA genotyping of Taenia solium. Korean J Parasitol 2011;49:399-403. PMID: 22355207.
17. Knapp J, Nakao M, Yanagida T, Okamoto M, Saarma U, Lavikainen A, Ito A. Phylogenetic relationships within Echinococcus and Taenia tapeworms (Cestoda: Taeniidae): an inference from nuclear protein-coding genes. Mol Phylogenet Evol 2011;61:628-638. PMID: 21907295.
18. Dai RS, Liu GH, Song HQ, Lin RQ, Yuan ZG, Li MW, Huang SY, Liu W, Zhu XQ. Sequence variability in two mitochondrial DNA regions and internal transcribed spacer among three cestodes infecting animals and humans from China. J Helminthol 2012;86:245-251. PMID: 21745429.
19. Varcasia A, Lightowlers MW, Cattoli G, Cancedda GM, Canu S, Garippa G, Scala A. Genetic variation within Taenia multiceps in Sardinia, Western Mediterranean (Italy). Parasitol Res 2006;99:622-626. PMID: 16614827.
20. Avcioglu H, Yildirim A, Duzlu O, Inci A, Terim KA, Balkaya I. Prevalence and molecular characterization of bovine coenurosis from Eastern Anatolian region of Turkey. Vet Parasitol 2011;176:59-64. PMID: 21074326.
21. Liu GH, Lin RQ, Li MW, Liu W, Liu Y, Yuan ZG, Song HQ, Zhao GH, Zhang KX, Zhu XQ. The complete mitochondrial genomes of three cestode species of Taenia infecting animals and humans. Mol Biol Rep 2011;38:2249-2256. PMID: 20922482.
22. Dai RS, Li F, Liu W, Lin RQ, He SW, Liu Y. Prevalence and identification of Coenurus on goats in Xiang-xi region. Chin J Zoonoses 2009;25:391-394 (in Chinese).
23. Jeon HK, Eom KS. Taenia asiatica and Taenia saginata: genetic divergence estimated from their mitochondrial genomes. Exp Parasitol 2006;113:58-61. PMID: 16546174.
Fig. 2Phylogenetic relationship among the examined Taenia species inferred by neighbor joining (NJ), maximum-parsimony (MP), and minimum-evolution (ME) analyses based on mitochondrial cox1 sequences, using Echinococcus granulosus as the outgroup. The numbers along branches indicate bootstrap values resulting from different analyses in the order: NJ/MP/ME. Values less than 50 are given as '-'. 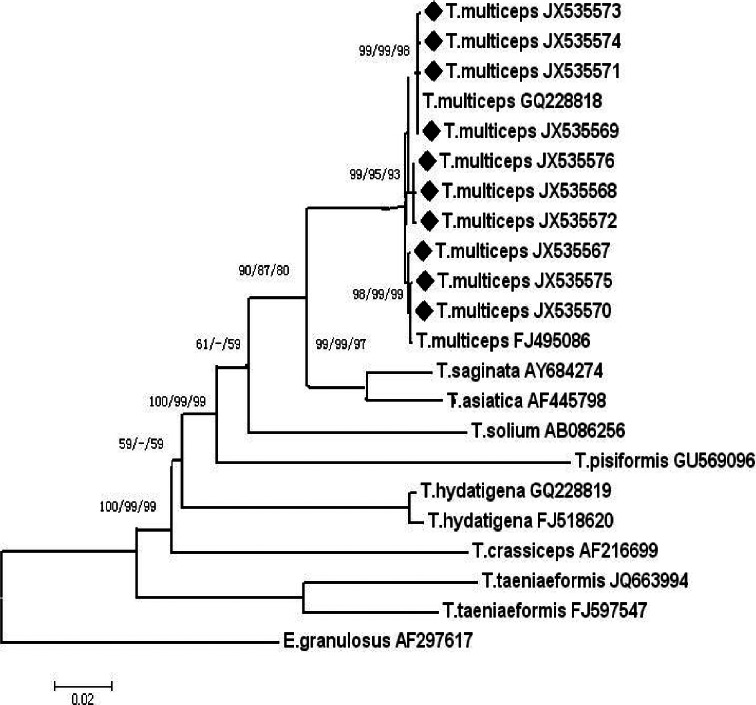
Table 1.
Taenia multiceps isolates used in the present study |
|