INTRODUCTION
Hookworms are common parasitic nematodes in the digestive tract of human beings, dogs and cats, which can cause a serious hazard to the host. The main hookworms infecting cats include Ancylostoma ceylanicum, A. tubaeforme, A. braziliense, and Uncinaria stenocephala. The recent investigations have shown that the cat’s hookworm infection in China is mainly caused by A. ceylanicum and A. tubaeforme [1,2]. A. ceylanicum is globally distributed among dogs and cats, however it is particularly severe in Asia, especially in Southeast Asia and the Pacific. Recently, A. ceylanicum has been recorded in many Southeast Asian countries, such as China [3,4], Japan [5], Malaysia [6], Laos [7], Thailand [8]. A. tubaeforme mainly infects cats, but it is rarely reported in canines. According to the previous surveys, A. tubaeforme has been reported in cats in Australia [9,10], Brazil [11], Italy [12], Qatar [13], Spain [14], the United States [15,16], and China [2]. Currently, A. ceylanicum has become the second largest species of hookworm in Asia [17], that can feed on blood and develop into adults in the human body, causing anemia, abdominal pain, diarrhea, and even death. As the number of cat owners increases, it is important to establish a rapid and accurate method that can distinguish A. ceylanicum from A. tubaeforme to enhance public health and safety.
The fecal examination is a classical method for hookworm detection, but it fails to identify hookworm species. Therefore, it is difficult to judge whether the infection is caused by A. ceylanicum. Presently, many molecular detection techniques have been applied to identify hookworms, such as specific polymerase chain reaction (PCR), multiplex PCR, restriction fragment length polymorphism (RFLP) and high-resolution melting (HRM) analysis [18–20]. All the former techniques have certain limitations, some can only detect a small number of samples at the same time, and others are complicated and expensive to operate.
Melting temperature shift (Tm-shift) method based on single nucleotide polymorphism (SNP) is a new molecular detection technique, which is high-throughput, accurate and inexpensive. This method utilizes 2 allele-specific primers, each of which attach to one of the two SNP loci at its 3′ end, a reverse primer that amplifies both alleles, and a fluorescent dye, SYBR® Green I (Molecular Probes, Eugene, Oregon, USA). Either one or both allele specific primer(s) amplify according the sample genotype. GC rich tails of unequal length are attached to the 5′ end of the allele-specific primers, so that the PCR product has a different melting temperature depending on which of the 2 primers is used. Therefore, genotypes can be distinguished by checking the melting curve on a real-time PCR instrument [21,22]. The Tm-shift method has been applied in many fields, such as medicine, microbiology, and aquatic biology [23–25]. In our laboratory, Pan et al. [26] established a Tm-shift genotyping method for Giardia lamblia assemblages A and F in cats, which pioneered a new genotyping method for veterinary parasitic protozoa. In addition, Fu et al. [27] developed a Tm-shift detection method for A. ceylanicum and A. caninum in dogs and applied it for the first time to identify veterinary parasitic nematode.
The present study aimed to develop and evaluate a Tm-shift method for the identification of A. ceylanicum and A. tubaeforme in cats, to provide a fast and accurate technique for the clinical detection and zoonotic risk assessment of cat-derived hookworms.
MATERIALS AND METHODS
Source of samples
Adult A. ceylanicum and A. tubaeforme samples were isolated and identified by Liu et al. [4] and Shi et al. [2], and preserved in 50% ethyl alcohol in our laboratory. Fecal samples were collected from stray cats at the Animal Rescue Station in Shaoguan city of Guangdong province in China and stored at 4°C.
Genomic DNA extraction
The preserved adult hookworms were repeatedly washed with double-distilled water (ddH2O). After that, the genomic DNA was extracted using the Wizard® SV Genomic DNA Purification System (Promega, Guangzhou, China) according to the manufacturer’s instructions, then stored at −20°C.
PCR amplification of ITS1 sequence
The extracted DNA was subjected to polymerase chain reaction targeting A. ceylanicum and A. tubaeforme internal transcribed spacer 1 (ITS1) sequences using a forward primer AF (5′-CTTTGTCGGGAAGGTTGG-3′) and a reverse primer AR (5′-TTCACCACTCTAAGCGTCT-3′) previously designed by Liu et al. [4]. The target amplification fragment length was 404 bp. PCR was carried out in a final volume of 25 μl, containing 12.5 μl Premix Ex-Taq polymerase (TaKaRa, Dalian, China), 9.5 μl ddH2O, 0.5 μl of each primer AF/AR (50 μmol/L), and 2 μl DNA template. The utilized PCR program was as follows: 94°C for 5 min, 35×(94°C for 30 sec, 61.5°C for 30 sec and 72°C for 45 sec), and 72°C for 7 min. The PCR products were assessed by electrophoresis in agarose gel (1.5%) with ethidium bromide stain (0.2 mg/ml) and visualized on a UV transilluminator. The amplified fragment was recovered from the gel following the protocol of the gel recovery kit (Omega, Guangzhou, China) and kept at −20°C until further use.
Preparation of standard plasmids
The purified PCR products were cloned in Escherichia coli and connected with pMD18-T vector (TaKaRa), then transformed into DH5 Competent Cells (TaKaRa). Positive clones were screened by bacterial PCR and sent to Sangon Biotech (Shanghai, China) for sequencing. The Plasmid Kit (Omega) was used to extract the plasmid DNAs. The plasmids harboring A. ceylanicum and A. tubaeforme ITS1 sequences were termed as AceP and AtuP. The plasmids (1 μl) were analyzed by the ultramicro-spectrophotometer to evaluate their purity and concentration, where A260/A280 value and optimal concentration should be 1.8–2.0 and 50 ng/μl, respectively.
Establishment of Tm-shift method based on SNP
Based on the 2 SNPs (ITS101 and ITS296), 2 sets of Tm-shift primers were designed using Primer Premier 5.0 software, referencing to the 2 sequences (MG733992, KP072069) downloaded from GenBank. The primer sequences and predicted amplification fragment length are demonstrated in Table 1. The above-mentioned primers were synthesized by Sangon Biotech (Shanghai, China). The synthesized primers were prepared in a template liquid at 100 pmol/μl and stored at −20°C. Then they were re-diluted to a final working concentration of 10 pmol/μl and stored at −20°C for use after partial shipments. PCR amplification and Tm-shift reaction were performed once in Rotor-Gene Q. The qPCR-Tm-shift reaction mixture had the final volume of 20 μl, containing 2×SYBR® Premix Ex Taq TM II (10 μl), long-tail primer (0.4 μl), short-tail primer (0.4 μl), common reverse primer (0.8 μl), ddH2O (7.4 μl), and plasmid (1.0 μl). The cycling program was as follows: 95°C for 5 min, 40×(95°C for 10 sec and 63°C for 30 sec). The melting process ranged from 70 to 95°C at the rate of 0.5°C/sec.
Stability, sensitivity and accuracy test
The stability of the established Tm-shift method was detected by repeating the experiments using 2 standard plasmids (AceP and AtuP). Intra-assay was checked 7 times, while inter-assay was checked 3 times for each plasmid. To evaluate the Tm-shift method sensitivity, AceP and AtuP were diluted 10 times and examined following the concentration of 1:10–1:108. To assess the Tm-shift method accuracy, 10 samples with known hookworm species were examined. The qPCR-Tm-shift reaction mixture and cycling program were identical to the mentioned above.
Clinical detection
Sixty-nine stray cat fecal samples were analyzed by zinc sulfate flotation technique and Tm-shift method. All fecal samples were suspended in ddH2O, heated for 5 cycles at 100°C for 5 min and immediately frozen at −80°C for 5 min. Genomic DNAs were extracted from the fecal samples using Stool DNA extraction kit (OMEGA, Guangzhou, China) as indicated by the manufacturer’s protocols. The qPCR-Tm-shift reaction mixture and cycling program were the same as the mentioned above. All results were further confirmed by DNA sequencing.
RESULTS
Amplification of ITS1 fragment and preparation of standard plasmids
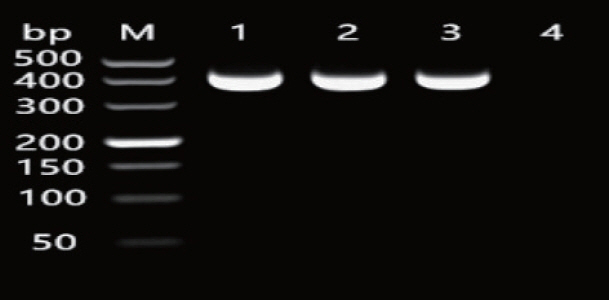
The amplification of the 2-hookworm species ITS1 sequences produced fragments with the length of 404 bp (Fig. 1), which agreed with the expected fragment length. The generated sequence data were submitted to GenBank (accession number: KF279132, JO812691). BLAST analysis showed the highest similarity (100%) with A. ceylanicum from Japan (LC036567) and the highest similarity (100%) with A. tubaeforme from Guangzhou (MG865903). Thus, 2 hookworms were identified as A. ceylanicum and A. tubaeforme. The A260/A280 values of positive plasmids of A. ceylanicum (AceP) and A. tubaeforme (AtuP) were between 1.8 and 2.0. In addition, the concentration of AceP and AtuP was 53.3 and 50.3 ng/μl, respectively, which was consistent with the optimal criteria.
Detection of qPCR-Tm-shift
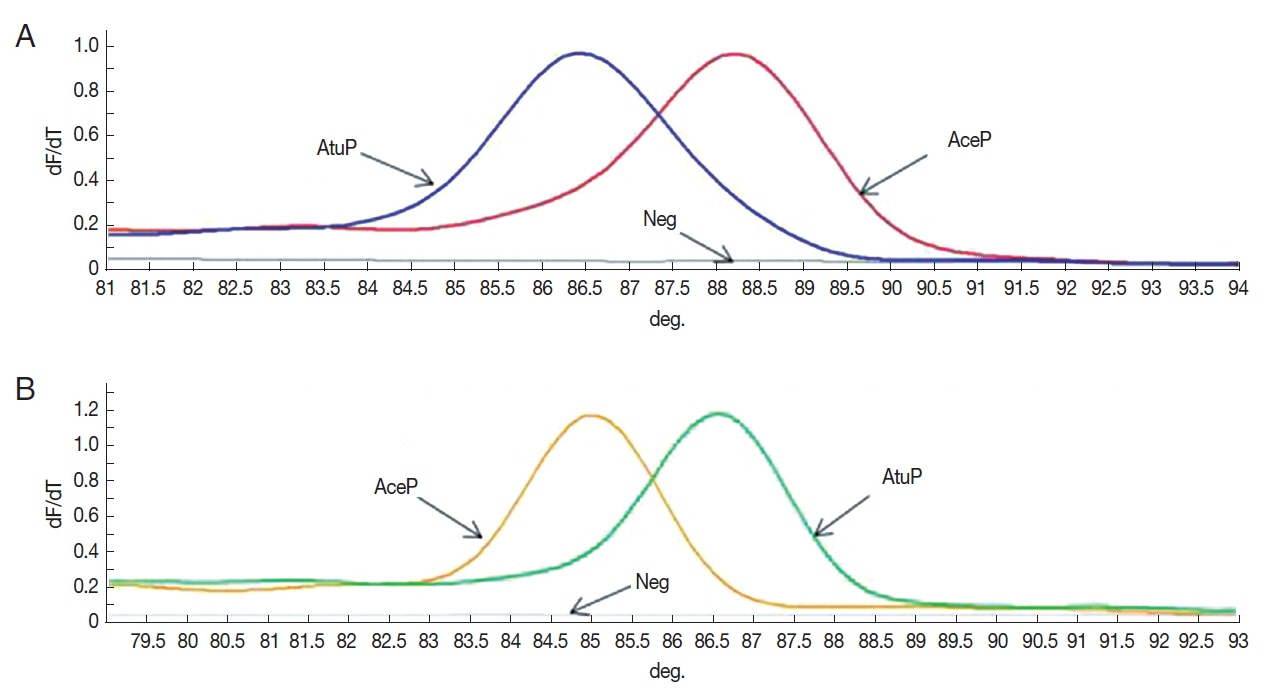
The Tm-shift standard curves based on the 2 SNPs (ITS101 and ITS296) for standard plasmid AceP and AtuP are revealed in Fig. 2. The result showed that the established Tm-shift method could distinguish A. ceylanicum (AceP) from A. tubaeforme (AtuP). The analysis of the standard curves by Rotor-Gene Q series software demonstrated that in the ITS101 primer, the Tm values of AceP and AtuP were 88.25°C and 86.50°C, respectively. In the ITS296 primer, the Tm values of AceP and AtuP were 85.00°C and 86.60°C, respectively.
Stability, sensitivity and accuracy
The stability test results are presented in Table 2. In the primer ITS101, the coefficient of variation (CV) of AceP and AtuP melting temperature (Tm) was 0.07% and 0.06%, respectively. In primer ITS296, the CV of AceP and AtuP melting temperature was 0.06% and 0.08%, respectively. The sensitivity test results indicated that the Tm-shift method could still differentiate between A. ceylanicum (AceP) and A. tubaeforme (AtuP), when they were diluted to 1:107 (5.22×10−6 ng/μl and 5.28×10−6 ng/μl, respectively) (Table 3). The results of examination of 10 DNA samples by Tm-shift method were identical to their known hookworm species with 100% accuracy (Table 4).
Clinical detection
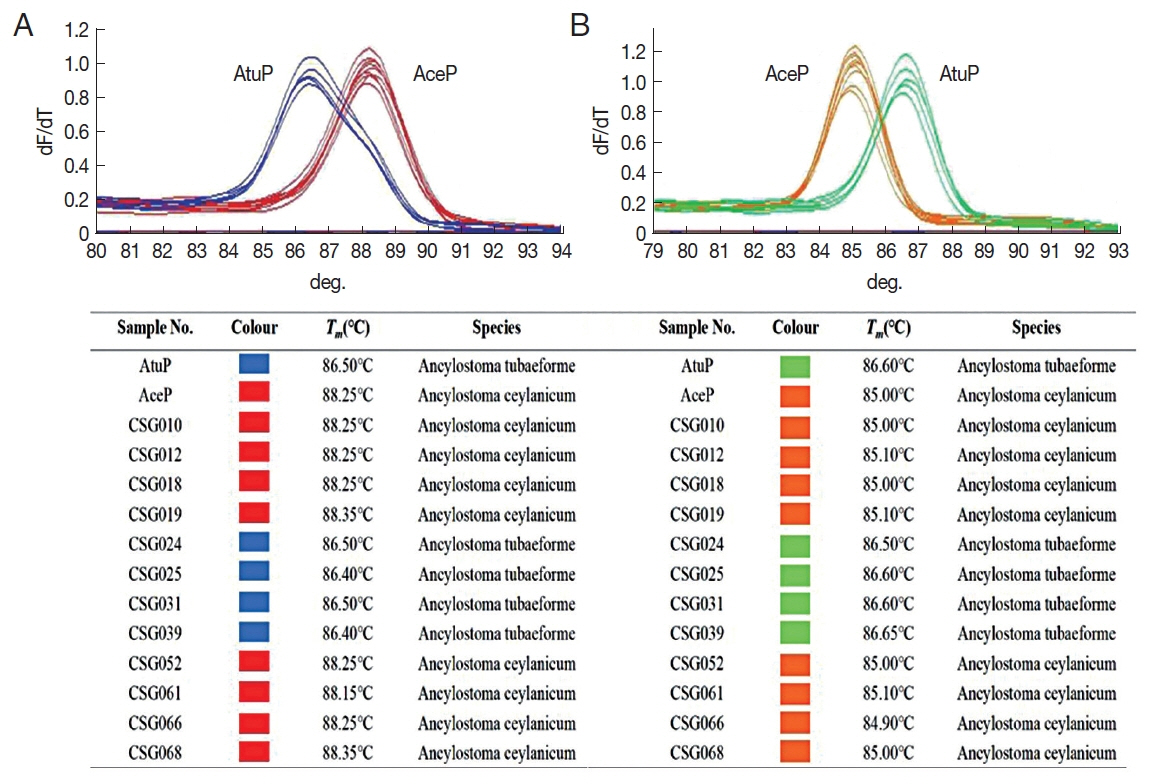
Twelve out of 69 cat fecal samples were positive for hookworm by the fecal flotation and Tm-shift methods. Using Tm-shift method, 8 and 4 samples were identified as A. ceylanicum and A. tubaeforme, respectively. The melting curves and detection results are shown in Fig. 3. All detection results were identical to the sequencing results.
DISCUSSION
Ancylostoma ceylanicum and A. tubaeforme are the 2 most common parasitic nematodes in cats. The former is the only animal-derived hookworm that can cause severe infection in humans. In Malaysia, the infection rate of A. ceylanicum is as high as 23.4%, which has become the second largest hookworm in local people [28]. In Laos, one-third of human hookworm infections are caused by A. ceylanicum [29]. Also, there are reports of human infection with A. ceylanicum in Taiwan [30]. In recent years, many cases of cat infection with A. tubaeforme have been detected in Guangzhou, China [2]. With the economic development and life quality improvement, more and more people are raising cats. Hence, it is of great significance to establish a Tm-shift detection method for cat-derived A. ceylanicum and A. tubaeforme for the prevention and control of zoonotic hookworm disease.
Common molecular biological methods for hookworm species identification include polymerase chain reaction (PCR), multiplex PCR, restriction fragment length polymorphism (RFLP), and high-resolution melting curve (HRM) [18–20]. Although the first 3 methods are specific and sensitive, they require gel electrophoresis after PCR, which makes the samples can be easily contaminated. In addition, the long-term use of ethidium bromide may seriously endanger the health of the operator. As a new molecular detection method, Tm-shift method is simple and quick to operate compared with the above-mentioned methods. The Tm-shift method is completed at one time on a closed fluorescent PCR instrument minimizing the contamination risk. As well as, it can examine 36 or 72 samples at the same time. Compared with HRM, the Tm-shift method does not require a relatively complicated HRM program. The used fluorescent dye in Tm-shift method is the ordinary SYBR Green I fluorescent dye, which is less expensive than the saturated dye required for HRM. Furthermore, the Tm-shift method can complete the amplification of the target fragment by using a 2-step method instead of 3 steps in HRM, which greatly saves the experiment time and improves the detection efficiency.
Even though the Tm-shift detection method has clear benefits over the other methods, it has some limitations during application. For example, the Tm-shift method has higher requirements for the design of 2 specific primers. The Tm value of the specific primer should be between 59°C and 62°C, and the primer length should be controlled in 15–22 bp. For a higher Taq polymerase amplification efficiency, the difference of Tm value between the reverse and specific primer (before adding sequences at the 5′ end) should be ranged from 4 to 5°C. In our study during the establishment of Tm-shift detection methods for cat-derived A. ceylanicum and A. tubaeforme, the primers were optimized where the short chain “gcggcg” was replaced with “gattaccg”. The results showed that this operation increased the Tm value of 2 melting curves to some extent. The difference between the peak of the melting curve for the 2 hookworms was 1.75°C at ITS101, while it was 1.60°C at ITS296. The established Tm-shift detection method was applied to examine 69 stray cat fecal samples in Shaoguan, Guangdong. The examination results of clinical fecal samples showed that 12 samples were positive for hookworm, from which 8 samples were A. ceylanicum and 4 samples were A. tubaeforme. The positive rate of Tm-shift method was consistent with the microscopic examination results, and this method could distinguish A. ceylanicum from A. tubaeforme. These results indicated that the designed Tm-shift method is rapid, accurate, efficient and sensitive.
It could be concluded that the developed Tm-shift method based on the 2 SNPs loci of ITS1 sequence can successfully identify A. ceylanicum and A. tubaeforme species. As well as, it can provide a suitable technical mean for clinical diagnosis and zoonotic risk judgment of cat-derived hookworms.