AbstractIn the present study, a Spirometra species of Tanzania origin obtained from an African leopard (Panthera pardus) and spotted hyena (Crocuta crocuta) was identified based on molecular analysis of cytochrome c oxidase I (cox1) and NADH dehydrogenase subunit I (nad1) as well as by morphological observations of an adult tapeworm. One strobila and several segments of a Spirometra species were obtained from the intestine of an African male leopard (Panthera pardus) and spotted hyena (Crocuta crocuta) in the Maswa Game Reserve of Tanzania. The morphological characteristics of S. theileri observed comprised 3 uterine loops on one side and 4 on the other side of the mid-line, a uterine pore situated posterior to the vagina and alternating irregularly either to the right or left of the latter, and vesicular seminis that were much smaller than other Spirometra species. Sequence differences in the cox1 and nad1 genes between S. theileri (Tanzania origin) and S. erinaceieuropaei were 10.1% (cox1) and 12.0% (nad1), while those of S. decipiens and S. ranarum were 9.6%, 9.8% (cox1) and 13.0%, 12.6% (nad1), respectively. The morphological features of the Tanzania-origin Spirometra specimens coincided with those of S. theileri, and the molecular data was also consistent with that of S. theileri, thereby demonstrating the distribution of S. theileri in Tanzania. This places the leopard (Panthera pardus) and spotted hyena (Crocuta crocuta) as new definitive hosts of this spirometrid tapeworm.
INTRODUCTIONCestode tapeworms of the genus Spirometra belong to the family Diphyllobothriidae which infect amphibians, birds and mammals. The adult stage of Spirometra spp. parasitize the intestine of carnivores, felids and canids including domestic dogs and cats. Spargana are plerocercoid larvae of cestode tapeworms of the genus Spirometra, which are found in paratenic hosts of Spirometra spp. such as amphibians, reptiles and mammals including humans.
Spirometrid tapeworms previously reported in African mammals were described by Baer (1924) [1]. S. pretoriensis was assigned the name Lühella pretoriense (Baer, 1924) and became synonymous with Diphyllobothrium pretoriense by Baer (1926), which was found in bat-eared fox (Otocyon megalotis) from South Africa (Baer, 1926) and later in African wild dog (Lycaon pictus) from Congo (Baer and Fain, 1955) [2]. S. theileri has been found in bush cat (Leptailurus serval), tiger cat (Felis lybica) near Pretoria, and lion from Congo (Baer, 1959) [3]. According to some studies, Spirometra and Taenia species were the most prevalent parasites in African lion, spotted hyena and African wild dog [4–8]. The spotted hyenas in Kenya and Zambia were reported to have a high rate of Spirometra infection with 74.3% and 22% in the feces of examined animals, respectively [5]. Some lions in Tanzania and Zambia were noted as having 63% and 87% infection of spirometrid tapeworms, respectively [5,6]. Spirometra species were the most commonly found in lion and second most common parasite in hyenas. The Spirometra species in lions is S. theileri whereas the species found in hyenas is generally accepted to be S. pretoriensis, although both Spirometra species may occur in lions [9]. The plerocercoids of Spirometra have been found in many herbivores including antelope, zebra and warthog in Kenya and Tanzania [8,9]. Baboons and other monkeys have also been infected with Spirometra species in East Africa [10,11]. Host susceptibility of African Spirometra species was reported from experimentally infected dogs and cat. They were able to infect dogs but not cats with spargana recovered from wart hogs in Tanzania [12]. They were also unable to infect various amphibians and reptiles with oral doses of infected copepods [12]. Although 2 Spirometra species have been reported as being S. theileri and S. pretoriensis from South Africa and Congo, it is unknown if these Spirometra species can infect humans. There have been a few reports and sporadic records of human sparganosis in Africa [13,14]. Human infections are unclear and not fully understand in Africa. Water containing procercoid larvae has been suggested as a possible mode of human infection in North and East Africa, and of wild animals in Masailand and Chad [13–15].
Previous studies have dealt with the validity of making Spirometra species assignments based on morphological characteristics that has given rise to 6 Spirometra species as independent species, 38 Spirometra species as the 1 species, and 4 species as valid species [16–18]. To date, the genus Spirometra includes S. erinaceieuropaei, S. decipiens, S. masonoides and S. ranarum as valid species [19–22]. In the present study, a Spirometra species of Tanzania origin obtained from an African leopard (Panthera pardus) and spotted hyena (Crocuta crocuta) was identified based on molecular analysis of cytochrome c oxidase I (cox1) and NADH dehydrogenase subunit I (nad1) as well as by morphological observations of an adult tapeworm.
MATERIALS AND METHODSSpecimens and morphological analysisOne strobila and several segments of a Spirometra species were obtained in intestine from an African male leopard (Panthera pardus) and spotted hyena (Crocuta crocuta) in the Maswa Game Reserve of Tanzania in February 2012. The Maswa Game Reserve (34°35′E, 03°16′S) was established in 1962, covers an area of 2,200 square kilometers, and is located in the south-western boundary of Serengeti National Park (34°50′E, 02°30′S). Parasite materials used in this study were provided by the Parasite Resource Bank of the Korea National Research Resource Center, Republic of Korea (PRB000720).
The collected worm was fixed in 70% ethanol and some gravid proglottids were compressed between 2 glass slides prior to fixation in alcohol-formalin-acetic acid (AFA) for carmine staining. Part of the specimen was subjected to hematoxylin and eosin (H-E) staining after longitudinal sections were obtained for observation of the cirrus pouch, cirrus sac, seminal vesicle, uterus and genital pore. The following morphological characteristics of the Spirometra species were compared to those of Baer [2] with carmine stained mature and gravid proglottids: width and length of mature and gravid proglottids, number of uterine loops, position of uterine pore, genital pore, vaginal opening, size and number of testes, and size of eggs.
PCR and DNA sequencingTotal genomic DNA extraction and PCR reactions were employed as previously described by Jeon et al. [21]. Mitochondrial cox1 and nad1 genes were amplified and sequenced by cycle sequencing. Primers used to amplify the mitochondrial cox1 gene (Spi-CO1F: 5′-GAC TAA GTG TTT TCA AAA CAC TAA GTG-3′ and Spi-CO1R: 5′CAC CCT ACC CCT GAT TTA CAA AAT-3′) and nad1 gene (Spi-ND1F: 5′-GGA GAA TAT TGG TTT GTC TAA CCA-3′ and Spi-ND1R: 5′-CCT TCT TAA CGT TAA CAG CAT TAC GAT-3′) were designed from the cox1 and nad1 sequences of S. erinaceieuropaei (KJ599680), S. decipiens (KJ599679) and S. ranarum (MH298843) mitochondrial genomes. PCR was performed in a 25 μl reaction volume containing 50 ng of genomic DNA, 10 pmol of each primer, 2.5 μl of 10× buffer, 12.25 μl of 2× buffer (MgCl2, dNTP) and 1.25 units Taq polymerase (TAKARA BIO INC, Kusatsu, Shiga, Japan). PCR was employed using a Biometra T-Gradient Thermoblock (Analytikjena, Göttingen, Germany) instrument under conditions comprising: 94°C for 3 min; 35 cycles of 94°C for 30 sec, 46°C for 40 sec, 72°C for 90 sec, and a final step at 72°C for 10 min. DNA sequencing was performed using a Big-Dye terminator kit (version 3.1, Applied Biosystems, Foster City, California, USA) and reaction products were sequenced directly using a DNA sequencer (ABI3730XL, Applied Biosystems).
DNA sequence analysesDNA sequences of the mitochondrial cox1 gene were assembled using the Geneious 9.1 program (Biometer, Auckland, New Zealand). These sequences were aligned using MAFFT methods in the Geneious 9.1 program by comparison with sequences of S. erinaceieuropaei, S. decipiens and S. ranarum in the GenBank database. Molecular Evolution Genetics Analysis (MEGA) software version 7.0 was employed to analyze nucleotide composition, conserved sites, variable sites, singleton sites and parsimony-informative sites [23]. Evaluation of the number of haplotypes, nucleotide diversity and haplotype diversity were performed using DnaSP software version 6.12 [24].
Phylogenetic analysis was evaluated by employing Bayesian inference (BI) and maximum-likelihood (ML) using the complete mitochondrial cox1 (1,566 bp) and nad1 (891 bp) sequences of S. erinaceieuropaei (KJ599680), S. decipiens (KJ599679), S. ranarum (MH298843, MH298844), Sparganum proliferum (AB015753), Dibothriocephalus nihonkaiense (=Diphyllobothrium nihonkaiense) (EF420138) and D. latus (=Diphyllobothrium latum) (DQ985706). The ML tree was constructed with 7 taxa and the nucleotide dataset of the mitochondrial cox1 gene as implemented in RAxML v. 7.3.1 [25] after TRN+G substitution model sampling was chosen according to the ModelTest using the program Partition Finder [26]. The nodes were assessed by bootstrapping with 1,000 pseudoreplicates. The BI tree was reconstructed with the same dataset using MrBayes 3.2 with a HKY+G substitution model and running 4 simultaneous Monte Carlo Markov chains (MCMC) for 10 million generations, sampling every 1,000 generations and discarding the first 25% of the generations as burn-in [27].
RESULTSMorphological characteristicsThe strobila collected from the intestine of a leopard (Panthera pardus) and spotted hyena (Crocuta crocuta) was 45 cm and 134.5 cm, respectively. The dimensions of immature to gravid proglottids ranged from 0.4 mm to 1.0 mm in width and 0.15 to 0.25 mm in length. The genital pore was situated ventrally on the midline in the anterior 1/5 of the segment. The vaginal pore was situated closely behind the male genital pore. The uterine pores were on the midline behind the anterior margin of the terminal ball 0.03 mm in diameter. The uterus consisted of 3–4.5 loops on each side. The dumbbell-shaped ovary was connected to the uterus and situated near the posterior margin of the segment (Fig. 1A, B). The longitudinal section of the cirrus sac showed an oval form, 0.16 mm in length and 0.13 mm in width. The seminal vesicle was elliptical and measured 0.13 in length and 0.14 mm in width (Fig. 1C) (Table 1). The testes measured about 0.06 mm in diameter. The eggs measured 0.054–0.059 mm in length and 0.033–0.035 mm in width.
Sequence divergence of mitochondrial cox1 and nad1 genesThe divergence of the mitochondrial cox1 and nad1 sequences of the Spirometra species collected from a leopard and spotted hyena (Tanzania origin) was 9.6%/13.0% (S. decipiens, KJ599670), 9.8%/12.6% (S. ranarum, MH298843), 10.1%/ 12.0% (S. erinaceieuropaei, KJ599680), 12.2%/− (Sparganum proliferum, AB015753), 16.7%/20.3% (D. latus, DQ985706) and 17.8%/21.8% (D. nihonkaiense, EF420138) (Table 2). The cox1 (1,566 bp) and nad1 (891) sequences of the Spirometra species including S. erinaceieuropaei, S. decipiens, S. ranarum and S. theileri revealed 227 and 157 polymorphic sites, 59 and 41 parsimony informative sites, and 169 and 116 singleton variable sites, respectively. There were 374.75 (cox1) and 209.00 (nad1) synonymous substitutions, and 1,188.25 (cox1) and 682.00 (nad1) non-synonymous substitutions. The genetic divergence of the cox1 and nad1 sequences of the Spirometra species ranged from 0 to 10.1% and 0 to 13.0%, respectively. These cox1 and nad1 genes showed high haplotype diversity (Hd) and low nucleotide diversity (Pi) (Table 3).
Phylogenetic analyses of the Spirometra species was conducted using 2 analytic methods (ML and BI) based on the cox1 genes of S. erinaceieuropaei, S. decipiens, S. ranarum, Sparganum proliferum, D. latus and D. nihonkaiense. The phylogenetic tree showed high level confidence values for 4 branches of the 4 Spirometra species that divided into 3 clades containing S. theileri, S. erinaceieuropaei and S. decipiens, and S. ranarum sub-clades (Fig. 2).
DISCUSSIONSpirometrid tapeworms found in African mammals were described as S. pretoriensis and S. theileri (under the name Diphyllobothrium theileri) by Baer (1924). The morphological characteristics of S. theileri described by Baer were as follows: the average length was 350 to 400 mm, and the greatest width was 3.3 mm. The length of the scolex ranged from 0.53 to 0.86 mm, and the width 0.25 to 0.3 mm. The first segments were 0.4 mm wide and 0.02 mm long, becoming 1.9 mm in length and 2.4 mm in width. The testes measured 0.11 to 0.15 vs 0.06 to 0.08 mm in spherical shape and occupied nearly all of the lateral fields of the medullary parenchyma. The cirrus measured 0.16 mm in length and 0.06 mm in diameter. The cirrus pouch measured 0.3 mm in length and 0.13 mm in diameter. The vaginal opening was situated behind the cirrus pouch. The ovary was situated on the dorsal side of uterus in the posterior region of the segment. The uterus was characterized by the great diameter of its coils, of which there were 3 on one side and 4 on the other side of the mid-line. The uterine pore was situated posterior to the vagina and usually alternating irregularly either to the right or left of the latter. The ova were thick-shelled and operculated, and measured 57×34 μm [2] (Baer, 1924).
Since then, Opuni and Muller (1974) reported on S. theileri adult tapeworms recovered from dogs that were fed plerocercoids obtained from a wart hog in Tanzania. The morphology of S. theileri that has been described included the following features: mature proglottids measured 8 to 12 mm in width with a spirally coiled uterus of 4 to 7 loops, the final centrally placed loops were much larger than the others and were constricted and hemispherical in shape [12]. Graber (1981) described this species in detail as possessing the following characteristics: mature or gravid proglottids measured 2.5–3.5 mm, the segments were wider than they were long and measured 0.24–0.36 mm in length in the anterior part and 2.5–3.5×1.2–1.3 mm in the middle and posterior part, with the last segment being a little narrow (2.4×1.95 mm), the scolex measured 0.6×0.18 mm and was elongated with a spoon-like shape, and the mean size of the eggs was 49×32.1 μm [28].
In the present study, the morphological features of the Spirometra species collected from the intestine of a leopard (Panthera pardus) and spotted hyena (Crocuta crocuta) were examined according to the aforementioned studies. The morphological features of 2 Spirometra species isolated from Tanzania resembled that of S. theileri in a previous study (Baer, 1926), whereby the uterus consisted of 3 loops on one side and 4 loops on the other side of the mid-line, and the aspect of the gravid uterus in the longitudinal section. The morphological characteristics of S. theileri have been described by Baer (1926) [2] as comprising a uterus with 3 uterine loops on one side and 4 on the other side of the mid-line, the uterine pore was situated posterior to the vagina and alternating irregularly either to the right or left of the latter, and the vesicular seminis were much smaller than other Spirometra species. The distinct features of S. theileri relative to other Spirometra species included a uterine morphology comprising uterine coils that were larger than the terminal ball and ellipse shape. The morphological features of S. theileri were distinct from that of S. pretoriensis. The major differentiating features concern the number of uterine loops and shape of the cirrus pouch. For example, the uterus of S. theileri consists of 3–4.5 coils and a cirrus pouch communicated through a short canal with a second, much smaller vesicular seminis, while the uterus of S. pretoriensis forms a single large loop on either side and the cirrus pouch possesses a large vesicular seminis of variable shape and size owing to the presence of very muscular walls [2].
Molecular identification of S. theileri was examined by sequence differences in the cox1 and nad1 genes with other Spirometra species including S. erinaceieuropaei, S. decipiens and S. ranarum. Sequence differences in the cox1 and nad1 genes between S. theileri (Tanzania origin) and S. erinaceieuroapei were 10.1% (cox1) and 12.0% (nad1), while those of S. decipiens and S. ranarum were 9.6% and 9.8% (cox1) and 13.0% and 12.6% (nad1), respectively. The nucleotide diversity (Pi) value of cox1 and nad1 was greater than 0.01 in this study, suggesting that the nucleotide sequences of Spirometra species such as S. erinaceieuropaei, S. decipiens and S. theileri have large variation. The phylogenetic analyses based on ML and BI methods suggested that the mitochondrial DNA sequences of S. theileri were separated from S. erinaceieuropaei and S. decipiens.
In conclusion, this study identified a Spirometra species of Tanzania origin by morphological examination and mitochondrial cox1 and nad1 sequences analysis of the adult worms. The morphological features and molecular data of the Tanzania-origin Spirometra specimens coincided with those of S. theileri, thereby demonstrating the distribution of S. theileri in Tanzania. S. theileri was found in leopard (Panthera pardus) and spotted hyena (Crocuta crocuta) as new definitive hosts of this spirometrid tapeworm in Tanzania.
ACKNOWLEDGMENTSThis work was supported by the National Research Foundation of Korea (NRF-2017K1A3A1A09085607). Materials used were provided by the Parasite Resource Bank of Korea (PRB 000720).
REFERENCES1. Baer JG. Contribution a la faune helminthologique sudafricaine. Note preliminaire. Annales de Parasitologie 1924;2:237-247 (in French).
2. Baer JG. Contribution to the helminth fauna of South Africa. Mammalian cestodes. In Baer JG ed, 11th and 12th Report of the Director of Veterinary Education and Research. Pretoria, South Africa. Department of Agriculture. 1926, pp 61-136.
3. Baer JG, Fain A. Helminthes parasites. Exploration des Parcs Nationaux du Congo Belge. Brussels, Belgium. 1959, p 163.
4. Engh AL, Nelson KG, Peebles R, Hernandez AD, Hubbard KK, Holekamp KE. Coprologic survey of parasites of spotted hyenas (Crocuta crocuta) in the Masai Mara National Reserve, Kenya. J Wildl Dis 2003;39:224-227.
5. Berensten AR, Becker MS, Stockdale-Walden H, Matandiko W, McRobb R, Dunbar MR. Survey of gastrointestinal parasite infection in African lion (Panthera leo), African wild dog (Lycaon pictus) and spotted hyaena (Crocuta crocuta) in the Luangwa Valley, Zambia. Afr Zool 2012;47:363-368.
6. Müller-Graf CD. A coprological survey of intestinal parasites of wild lions (Panthera leo) in the Serengeti and the Ngorongoro Crater, Tanzania, East Africa. J Parasitol 1995;8:812-814.
7. Bjork KE, Averbeck AG, Stromberg BE. Parasites and parasite stages of free-ranging wild lions (Panthera leo) of northern Tanzania. J Zoo Wild Med 2000;31:56-61.
8. Müller-Graf CD, Woolhouse ME, Packer C. Epidemiology of an intestinal parasite (Spirometra spp.) in two populations of African lions (Panthera leo). Parasitology 1999;118:407-415.
9. Nelson GS, Pester FRN, Rickman R. The significance of wild animals in the transmission of cestodes of medical importance in Kenya. Trans R Soc Trop Med Hyg 1965;59:507-523.
10. Kuntz RE, Myers BJ, Katzberg AA. Sparganosis and “proliferative” spargana in vervets (Cercopithecus aethiops) and baboons (Papio sp.) from East Africa. J Parasitol 1970;56:196-197.
11. Nobrega-Lee M, Hubbard G, Loverde P, Carvalho-Queiroz C, Conn DB, Rohde K, Dick EJ Jr, Nathanielsz P, Martin D, Siler-Khodr T, Schlabritz-Loutsevitch N. Sparganosis in wild-caught baboons (Papio cynocephalus anubis). J Med Primatol 2007;36:47-54.
12. Opuni EK, Muller RL. Studies on Spirometra theileri (Baer, 1925) n. comb. 1. Identification and biology in the laboratory. J Helminthol 1974;48:15-23.
13. Kavana NJ, Kassuku AA. Studies on community knowledge, behavior and practice on sparganosis infection in Mto wa Mbu, Monduli District, Tanzania. Basic Res J Med Clin Sci 2015;4:125-129.
14. Kavanas N, Sonaimuthu P, Kasanga C, Kassuku A, Al-Mekhafi HM, Fong MY, Khan MB, Mahmud R, Lau YL. Seroprevalence of sparganosis in rural communities of Northern Tanzania. Am J Trop Med Hyg 2016;95:874-876.
15. Eberhard ML, Thiele EA, Yembo GE, Yibi MS, Cama VA, Ruiz-Tiben E. Case report: thirty-seven human cases of sparganosis from Ethiopia and South Sudan caused by Spirometra spp. Am J Trop Med Hyg 2015;93:350-355.
16. Faust EC, Campbell HE, Kellogg CR. Morphological and biological studies on the species of Diphyllobothrium in China. Am J Epidemiol 1929;9:560-583.
17. Yamaguti S. Systema helminthum. II:The cestodes of vertebrates. New York, USA. Interscience Publishers. 1959, pp 358-361.
18. Kamo H. Guide to identification of diphyllobothriid cestodes. Gendai Kikaku. Tokyo, Japan. 1999, pp 1-146 (in Japanese).
19. Mueller JF. New host records for Diphyllobothrium mansonoides Mueller 1935. J Parasitol 1937;23:313-315.
20. Jeon HK, Park HS, Lee DM, Choe SJ, Kim KH, Huh S, Sohn WM, Chai JY, Eom KS. Human infections with Spirometra decipiens plerocercoids identified by morphologic and genetic analyses in Korea. Korean J Parasitol 2015;53:299-305.
21. Jeon HK, Park HS, Lee DM, Choe SJ, Kang YS, Bia MM, Lee SH, Sohn WM, Hong SJ, Chai JY, Eom KS. Genetic and morphologic identification of Spirometra ranarum in Myanmar. Korean J Parasitol 2018;56:275-280.
22. Eom KS, Park HS, Lee DM, Choe SJ, Kang YS, Bia MM, Lee SH, Keyyu J, Fymagwa R, Jeon HK. Molecular and morphologic identification of Spirometra ranarum found in the stool of African lion, Panthera leo in the Serengeti plain of Tanzania. Korean J Parasitol 2018;56:379-383.
23. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA7 molecular evolutionary genetics analysis using maximum likelihood evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;25:2731-2739.
24. Librado P, Rozas J. DnaSP6: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009;25:1451-1452.
25. Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006;22:2688-2690.
26. Lanfear R, Calcott B, Hos YW, Guindon S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 2012;29:1695-1701.
Fig. 1Gravid proglottids of Spirometra theileri (Tanzania origin) collected from a leopard (Panthera pardus) (A) and spotted hyena (Crocuta crocuta) (B). (A, B) Whole mounted specimens of proglottids showing the genital pore (GP), vaginal pore (VP), and uterus (U) (×40), (C) Longitudinal section of a gravid proglottid showing the cirrus sac (CS), seminal vesicle (SV), and uterus (U) (H&E stain). 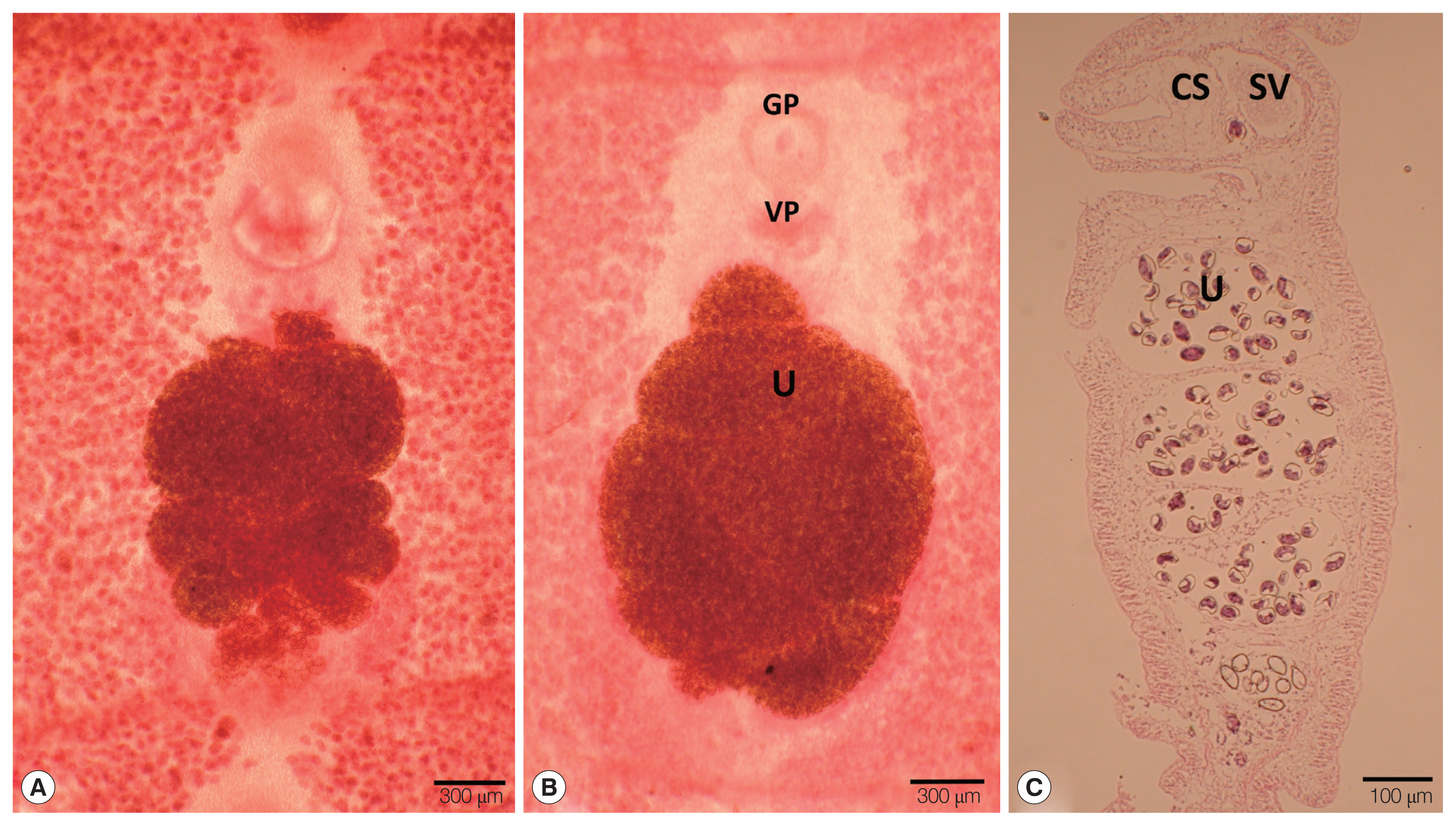
Fig. 2Phylogenetic relationship of Spirometra species based on cox1 and nad1 sequences. Numbers on branches represent bootstrap values for maximum likelihood (ML) and the support value of Bayesian inference (BI). GenBank No. MK955901 and MN244299 are registered as S. theileri by complete mitochondrial cox1 and nad1 sequences. 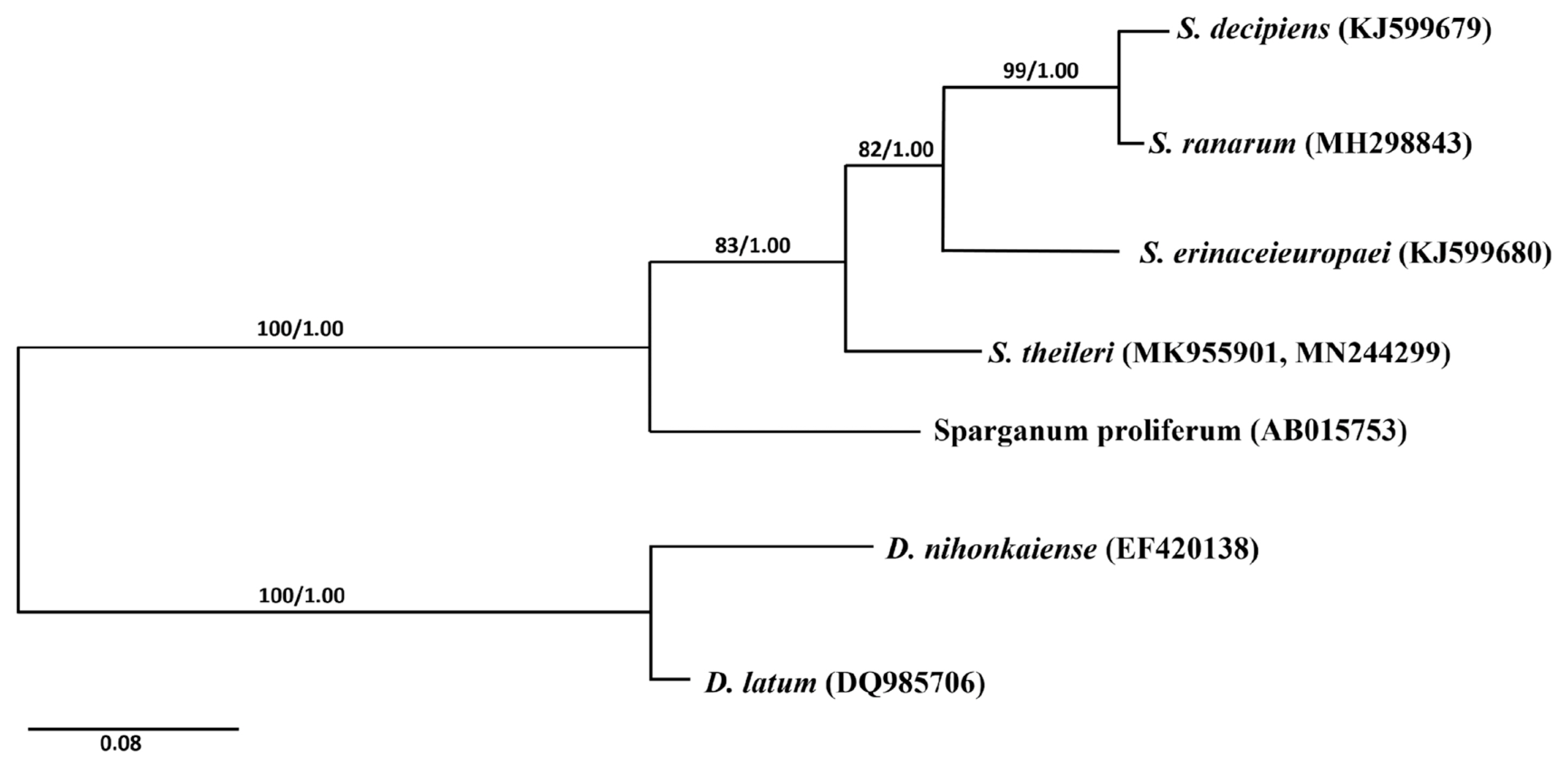
Table 1Morphological features of Spirometra theileri and Spirometra sp.
Table 2Pairwise sequence differences of mitochondrial cox1 and nad1 genes of Spirometra species of Tanzania origin and various Spirometra species, Dibothriocephalus latus and D. nihonkaiense
Table 3Genetic diversity of mitochondrial DNA sequences of Spirometra species including S. erinaceieuropaei, S. decipiens, S. ranarum and S. theileri
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||