Phylogenetic Relationships of 3 Korean Neodiplostomum Species (Digenea: Neodiplostomidae) Based on Partial CO1 Gene
Article information
Abstract
The phylogenetic relationships of the 3 Neodiplostomum spp. (Digenea: Neodiplostomidae) occurring in Korea (N. seoulense, N. leei, and N. boryongense) were analyzed using the partial mitochondrial cytochrome c oxidase subunit 1 (CO1) gene. The adult flukes were recovered from Sprague-Dawley rats (N. seoulense) and newborn chicks (N. leei and N. boryongense) experimentally infected with the neodiplostomula from the grass snake, Rhabdophis tigrinus tigrinus. The genomic DNA was amplified using specific primers, and the sequence of CO1 was obtained. According to the results, the pairwise similarity was 96.1% between N. boryongense and N. seoulense, but was 95.0% between N. boryongense and N. leei and 94.2% between N. leei and N. seoulense. The results demonstrated a closer phylogenetic relationship between N. seoulense and N. boryongense. This high relationship of N. seoulense and N. boryongense may be related to their similar morphologic features including the limited distribution of vitellaria and the presence of a genital cone. N. leei is distinct on the other hand with an extensive distribution of vitellaria and the absence of a genital cone.
The grass snake, Rhabdophis tigrinus tigrinus (Boie, 1826), inhabiting the Republic of Korea, has at least 3 species of small-sized neodiplostomula (metacercariae of neodiplostomid flukes), i.e., Neodiplostomum seoulense, Neodiplostomum leei, and Neodiplostomum boryongense [1,2,3,4]. These neodiplostomula are indistinguishable based on their morphology; however, the definitive hosts and life cycles are different. N. seoulense develops into an adult fluke in the small intestine of rodents, as well as humans, whereas N. leei and N. boryongense develop in the small intestine of chicks but not in rodents [2,3]. The metacercariae of N. leei, when given orally to mice and rats, migrate to the liver of these rodent hosts. On the other hand, N. seoulense and N. boryongense do not migrate to visceral organs of rodent hosts [2,3]. Moreover, these neodiplostomula have a different destiny in the murine small intestine; N. seoulense develops into adult flukes, whereas N. boryongense do not develop into adults, but disappear from the intestine in a few days after infection.
Considering their common second intermediate (or paratenic) host with similar geographical distribution and similar morphologies of metacercariae and even adults, the 3 species of Neodiplostomum may have a close phylogenetic relationship. To analyze the degree of relationships between different parasite species, partial mitochondrial genes have been used [5]. In particular, the mitochondrial cytochrome c oxidase subunit 1 (CO1) gene has been used for DNA-based identification of parasites, providing the resolution of diversity based on the conserved mitochondrial gene [6]. The examples of parasites studied include Echinococcus spp. [7], Taenia multiceps [8], Taenia taeniaeformis, and other taeniid cestodes [9], Echinostoma species [10], Plagiorchis spp. [11], Ancylostoma duodenale and Necator americanus [12], Spirometra and Diphllobothrium [13], and rodent pinworms [14]. Although ITS [15] is also used for taxonomic studies, CO1 gene has been suggested to be useful for general evolutionary studies [16]. The present study was performed to analyze the phylogenetic relationships of the 3 species of Neodiplostomum occurring in Korea, using the partial CO1 gene.
Newborn broilers were purchased from a hatchery in Gyeonggi-do, Korea. Female Sprague-Dawley rats, 6-8-week-old, which were raised under specific pathogen-free (SPF) conditions, were purchased from Samtako (Osan, Gyeonggi-do, Korea). The grass snakes, R. tigrinus tigrinus, naturally infected with the neodiplostomula, were purchased from local snake collectors in enzootic areas, such as Osan and Ansung in Gyeonggi-do (Province), Hongcheon and Hoeongsung in Gangwon-do, and Cheonan, Onyang, Hongsung, and Boryong in Chungchongnam-do. Snakes were kept in a cold room for hibernation until used.
Small-sized neodiplostomula were collected by the digestion technique using artificial gastric juice containing 0.5% pepsin (1:10,000, Sigma-Aldrich, St. Louis, Missouri, USA) and 0.8% HCl. Snakes were sacrificed under ether anesthesia, and skinned [3]. The entire visceral mass and carcasses were separated and chopped before incubation in pepsin-HCl solution. The digested mixture was incubated for 90 min at 37℃ and washed several times by filtering through coarse and fine stainless steel meshes (US standard testing sieves, 425 µm, 300 µm, 150 µm, and 106 µm, W. S. Tyler Incorp., Mentor, Ohio, USA). After final filtration, the mixture was transferred to a Petri dish, and neodiplostomula were collected using a stereomicroscope. The whole procedure was carried out under cold conditions to maintain the viability of the neodiplostomula. Large-sized diplostomula of Pharyngostomum cordatum were carefully collected and removed [3].
Rats were orally infected with the neodiplostomula using a gavage needle and adult flukes of N. seoulense were collected from the small intestine at week 2 post-infection (PI). For collection of N. leei adults, metacercariae which were recovered from the liver of rats infected with small-sized neodiplostomula, were orally infected to newborn chicks, and adult flukes were recovered from the intestine of chicks at week 2 PI. The procedure of harvesting the neodiplostomula from the rat liver was as described previously [17]. The livers of infected rats were resected, cut into small pieces, digested with artificial gastric juice, and then the neodiplostomula were collected under a stereomicroscope. For collection of N. boryongense adult flukes, small-sized neodiplostomula recovered from snakes were directly fed to newborn chicks and the adult flukes were recovered from the intestine of chicks at week 2 PI. To collect the adult flukes, the intestine was taken out and put on a Petri dish containing saline, and then opened longitudinally. Adult flukes were collected under a stereomicroscope.
For cloning of CO1 gene from the genomic DNA of Neodiplostomum species, the primer set was prepared as follows [10]: the forward primer 5'-TTT TTT GGG CAT CCT GAG GTT TAT-3' and the reverse primer 5'-TCA TGA AAA CAC CTT AAT ACC-3'. The genomic DNA of neodiplostomid flukes was individually extracted using the DNA-EASY kit (Invitrogen, Carlsbad, California, USA) according to the manufacturer's instructions. DNA was amplified with CO1 specific primers and PCR premix (Bioneer, Seoul, Korea). Amplification was performed in a gradient Master cycler (Eppendorf, New York, USA) under the following conditions: 30 cycles consisting of 1 min at 94℃, 1 min at 49℃, and 1 min at 72°. Unincorporated primers and nucleotides were removed from the PCR product using the QIA quick gel extraction kit (Qiagen, Hilden, Germany). PCR products were ligated into a TA cloning vector in TOPO TA cloning kit (Invitrogen) and the subcloned DNA was sequenced (Cosmo Genetech Inc., Seoul, Korea) and analyzed using a bioinformatics tool (Geneious Pro 5.4.7, Geneious Co., Wellington, New Zealand). Because CO1 gene products were of different nucleotide sizes, the overlapped CO1 gene domain among the 3 species of Neodiplostomum flukes were used for this phylogenetic analysis.
We obtained partial CO1 gene sequences of the 3 species of neodiplostomid flukes (Fig. 1). An analysis program (Geneious Pro 5.4.7; Geneious Co.) was used for analysis of the gene alignment and phylogenetic tree. The partial CO1 gene of Fasciola hepatica (GenBank accession no. AB207170.1) was used as an outgroup. For alignment of the 3 CO1 genes, the sequenced DNA was processed by the Geneious Pro 4.6.2 program (Geneious Co.). The results were as follows: cost matrix was the lowest among the list with 51% similarity, gap open penalty score was 12, gap extension penalty score was 3, and alignment type was global alignment in general. The partial CO1 gene sequences studied were N. boryongense (280 bp), N. leei (276 bp), and N. seoulense (283 bp).

Partial cytochrome c oxidase subunit 1 (CO1) sequences of 3 Neodiplostomum species. Dash indicates gap introduced into alignments among the 3 trematode parasites.
According to the results, the pairwise similarity between N. boryongense and N. leei was 95%, and the pairwise similarity between N. leei and N. seoulense was 94.2%; the identical sites were 270 bp (93.4%) and 268 bp (94.4%) respectively. On the other hand, the pairwise similarity between N. boryongense and N. seoulense was 96.1% and the identical sites were 272 bp (94.1%). The genetic distance model used was 'Tamura-Nei', and the tree-built method was 'Neighbor-joining'. The results showed that the score using "PATRISTIC DISTANCES MATRIX" between N. boryongense and N. leei was 0.993, that between N. seoulense and N. leei was 0.919, and that between N. seoulense and N. boryongense was 0.073. Therefore, N. seoulense and N. boryongense were genetically closer than they were with N. leei (Fig. 2).
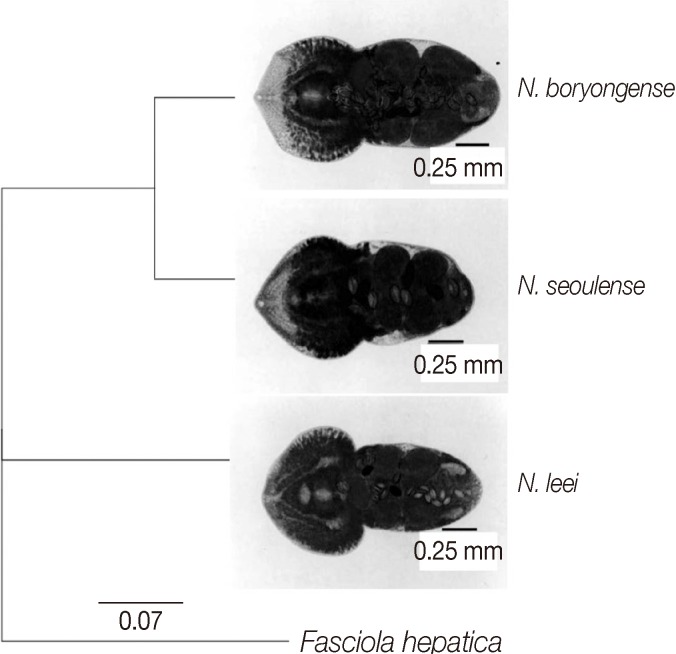
Phylogenetic tree inferred from the aligned sequence of partial CO1 gene. The sequence of Fasciola hepatica (GenBank accesion no. AB207170.1) as an outgroup, and sequences of Neodiplostomum spp. (N. boryongense, N. seoulense, and N. leei) were compared and aligned using the Geneious Pro 5.4.7 program. The genetic distance represents the dissimilarity among the 4 kinds of nucleotide sequences.
The present study revealed that the genetic proximity between N. seoulense and N. boryongense was higher than that between the other 2 combinations. The sequence similarity of CO1 gene was 96.1% between N. seoulense and N. boryongense, whereas that between N. leei and N. seoulense was 94.2% and that between N. leei and N. boryongense was 95.0%. It was highly suggested that all these 3 Neodiplostomum spp. have a common ancestry, and N. seoulense and N. boryongense may have differentiated later than N. leei. Neodiplostomid flukes may have progressed to be adapted from birds to mammals through predator and prey interactions [5,18]. Therefore, N. seoulense may be the newest clade followed by N. leei and N. boryongense.
The closer genetic relationship between N. seoulense and N. boryongense compared to N. leei may be phenotypically related to the morphology and life cycles of these neodiplostomid flukes. N. seoulense has vitellaria which are less extensive than N. leei distributing only up to the level of the ventral sucker, and 2 bilobed testes resembling a butterfly-shaped cuff link [4]. N. leei has vitellaria which are very extensive and distributed from the pharynx to near the posterior end of the body; it has no genital cone but has 2 bilobed testes with a long, prominent median constriction forming a dumbbell shape [2]. N. boryongense has vitellaria which are not extensive and distributed chiefly in the forebody without distribution between the pharynx and the ventral sucker [3]. Their testes are severely bilobed and eye-glass shaped.
The genital cone has taxonomic significance and is used to differentiate the genus Neodiplostomum into 2 subgenera, Neodiplostomum (Conodiplostomum) Dubois, 1937 and Neodiplostomum (Neodiplostomum) Dubois, 1937 [2,3]. N. seoulense and N. boryongense, which belong to Neodiplostomum (Conodiplostomum), have a genital cone near the terminal portion of the genital opening, whereas N. leei has no genital cone and thus belongs to Neodiplostomum (Neodiplostomum) [2,3]. Moreover, N. leei uses rodents as a paratenic host in which its neodiplostomula migrate to the liver [2]. However, N. seoulense and N. boryongense do not migrate to the liver of rodents and develop into adults in the intestine of rodents and chicks, respectively [2,3]. These comparative characters support our results of closer genetic proximity between N. seoulense and N. boryongense than with N. leei.
The phylogenetic analysis of only cox1 gene may not be sufficient to properly explain overall coevolution of parasites and their hosts [5]. However, our results could provide at least some knowledge on genetic proximity among the 3 neodiplostomid flukes occurring in Korea. Further biological and genetic studies will be helpful to support our results.
ACKNOWLEDGMENTS
We thank Mr. Jae-Lip Kim for his help in recovery of Neodiplostomum spp. adults from experimental rats and chicks. This study was supported by a grant from Seoul National University Hospital Research Fund (04-2009-0940).
Notes
We have no competing interest related to this work.