Toxoplasmosis is caused by the zoonotic parasite Toxoplasma gondii—a member of the order Coccidia and phylum Apicomplexa. Toxoplasma gondii is one of the most prevalent apicomplexan parasites of humans and other warm-blooded animals [1], infecting approximately one-third of the human population worldwide. The tissue cysts of T. gondii are frequently observed in goats, pigs, and lambs, which are food-producing animals. By contrast, cattle are a poor host for T. gondii, and tissue cysts are rarely found in beef [2]. Considering that the parasite leaves the bloodstream and forms a cyst in the muscle, cattle play a unique role in the epidemiology of toxoplasmosis. Infections in humans may be transmitted through the consumption of undercooked meat and milk [3]. Recently, the European Food Safety Authority evaluated the risk hierarchy associated with the consumption of raw or undercooked meat and concluded that beef poses a greater health risk than previously understood [4].
The consumption of raw cattle meat is a part of the cultural and traditional history of several countries as well as characterizes the current dietary trends. In Korea, beef is associated with a potentially high risk of transmitting the infection owing to consumer habits; compared with the meat of other animal species, beef is more frequently consumed raw or undercooked. These habits significantly affect public health [2]. Several seroprevalence studies regarding T. gondii in cattle have been conducted in Korea [5–7]. However, to date, information regarding the molecular epidemiology of T. gondii in cattle in Korea is lacking. Therefore, the number of cattle infected, location of endemic hotspots, percentage of infected beef present in food markets, and role of contaminated meat in the overall epidemiology of T. gondii remain unknown. Considering the lack of information on the molecular detection of T. gondii in cattle destined for human consumption, in the present study, we aimed to genetically characterize the infectious strains of T. gondii.
The Ministry of Agriculture, Food, and Rural Affairs in Korea conducts annual monitoring activities for contagious diseases on outdoor farms through the national livestock health program under the regulation of the “Act on the Prevention of Contagious Animal Disease (amendment Act 2017)”. Typically, blood samples are collected from animals by practicing veterinarians at official local veterinary government institutes, with verbal consent being obtained from the animal owners during monitoring, surveillance, treatment, and routine checkups. The cattle blood samples used in the present study were obtained from the annual bovine tuberculosis surveillance program, conducted for national livestock disease control purposes. Because these samples were used to confirm the presence of T. gondii infection in this study, approval from the Institutional Animal Care and Use Committee at Kyungpook National University was not required.
In 2017, overall, 3,428,330 cattle were reared in 105,073 farms in Korea [8]. Among them, 354,780 (10.3%) cattle were reared on 14,622 (13.9%) farms in the Gyeongnam Province. The sample size was determined using a formula with an expected disease prevalence of 10%, an accepted absolute error of 5%, and a confidence level of 95% with simple random sampling design [9]. According to the formula, a minimum of 138 samples was required. We randomly selected 455 cattle from 84 farms in the Gyeongnam Province during monitoring or surveillance in 2017. Data on age, sex, and breed were recorded for subsequent analysis.
Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Nested PCR was performed using the AccuPower HotStart PCR Premix Kit (Bioneer, Daejeon, Korea). To detect T. gondii, nested PCR was performed by amplifying the B1 gene, as previously described [10]; external (Tg1 and Tg2) and internal (Tg3 and Tg4) primers were used that generated a 531-bp amplicon. A sample of T. gondii isolated from cats in Korea [11] was used as the positive control, whereas a sample lacking a DNA template was used as the negative control.
The amplicons from the infected animals were purified using the QIAquick Gel Extraction Kit (Qiagen), ligated into the pGEM-T Easy vector (Promega, Madison, WI, USA), and transformed into Escherichia coli DH5α-competent cells (Thermo Fisher Scientific, Wilmington, DE, USA). Plasmid DNA extraction was performed using a plasmid miniprep kit (Qiagen) according to the manufacturer’s instructions.
Recombinant clones were selected and sent to Macrogen Inc., Daejeon, Korea, for nucleotide sequencing. The sequences obtained in this study were aligned and analyzed using the CLUSTAL Omega multiple sequence alignment program Omega v.1.2.1 (Bioweb, Ferndale, WA, USA); the alignment was corrected using the BioEdit software v.7.2.5 (Bioedit Company, Manchester, UK). Subsequently, phylogenetic analysis was performed using the MEGA software v.7.0 (Mega Software Solutions, Madhurawadha, India) based on the maximum likelihood method and employing the Kimura 2-parameter distance model. The aligned sequences were analyzed using a similarity matrix. The stability of the trees obtained was estimated using bootstrap analysis with 1,000 replicates.
The GraphPad Prism software v.5.04 (GraphPad Software Inc., La Jolla, CA, USA) was used for statistical analyses. A P-value of <0.05 was considered statistically significant. A 95% confidence interval (CI) was calculated for all estimates.
Among the 455 cattle from 84 farms, 3 cattle (0.7%, 95% CI: 0–1.4) from 3 farms (3.6%, 95% CI: 0–7.5) tested positive for the T. gondii B1 gene (Table 1). The prevalence of T. gondii was significantly higher in male cattle (4.2%, 2/48, 95% CI: 0–9.8) than in female cattle (0.2%, 1/407, 95% CI: 0–0.7; P=0.0015). With regard to age, T. gondii was the most prevalent in cattle aged >6 years (0.9%, 1/109, 95% CI: 0–2.7), followed by those aged 4–6 years (0.7%, 1/143, 95% CI: 0–2.1) and 1–3 years (0.6%, 1/167, 95% CI: 0–1.8); no prevalence was detected in cattle aged <1 year. The prevalence of T. gondii tended to be higher in older cattle than in young cattle; however, no significant differences were observed (P=0.9479). Regarding breed, T. gondii was only detected in brown cattle (0.9%, 3/333, 95% CI: 0–1.9) and not in dairy cattle; no significant difference was observed (P=0.2934). With regard to farm size, T. gondii was only detected in small farms (≤20 animals, 1.9%, 3/156, 95% CI: 0–4.1) and not in large farms (>20 animals).
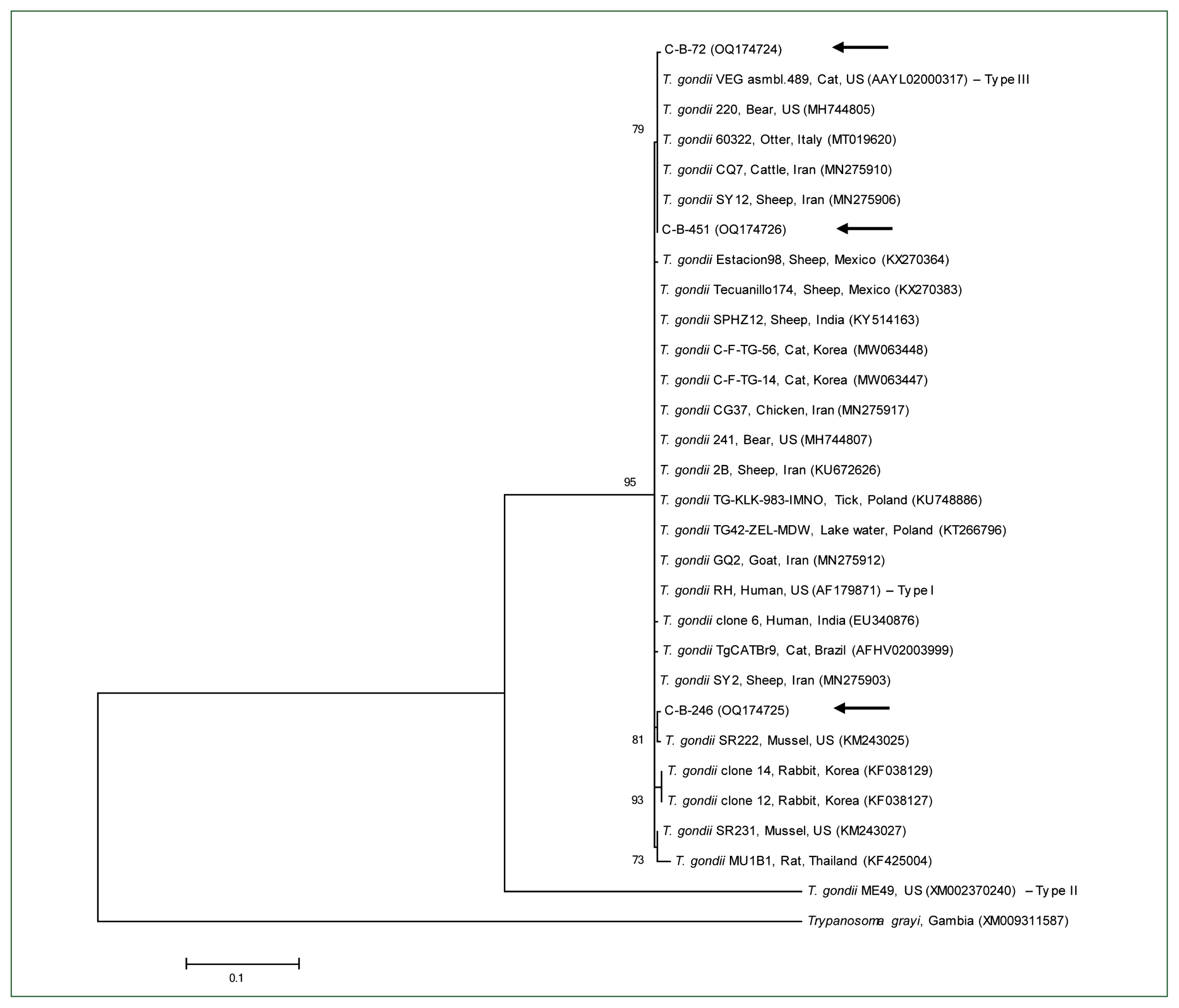
In phylogenetic analysis, the B1 gene nucleotide sequences of T. gondii identified in this study were found to be clustered with previously known T. gondii sequences (Fig. 1). The 3 B1 gene sequences of T. gondii in this study shared 99.3%–99.8% identity. In addition, they were 97.3%–99.6% identical to the B1 gene sequences of previously reported T. gondii isolates. All sequences used in the phylogenetic analysis were submitted to the GenBank database (accession numbers: OQ174724–OQ174726).
Given the significance of the meat industry worldwide, T. gondii infection in meat-producing cattle can result in destructive consequences on the economy as well as human health [12]. Undercooked beef is considered to be a source of T. gondii transmission to humans, particularly in regions where raw, undercooked, or cured beef is commonly consumed [4]. Several studies have been conducted regarding T. gondii infection in cattle. In Korea, T. gondii was detected in cattle in the Gyeongbuk Province using ELISA (0.5%, 3/568) in 2013 [5], in Ulleung Island using ELISA (17.6%, 97/552) between 2007–2010 [7], and in the eastern areas of the Gyeongbuk Province using ELISA (20.7%, 76/368) in 2008 [6]. Moreover, in Poland, T. gondii was detected in cattle using ELISA (13%, 313/2411) and in tissues using PCR (10.2%, 32/313) between 2009 and 2013 [13]; in Iran, it was detected in tissues using PCR (56%, 70/125) in 2017 [14]. Blood PCR is typically more beneficial for the early and late diagnosis of toxoplasmosis [15]; however, only 0.7% of cattle in the present study were positive for toxoplasmosis. It is possible that T. gondii possesses a low sensitivity to blood and that different parasite strains and infection pathways result in different blood parasitemia symptoms in primary infections [15]. In the present study, the low prevalence of T. gondii infection in cattle can be attributed to their high natural resistance to the parasite. Toxoplasma cysts can be eliminated, resulting in subclinical T. gondii infection in cattle [16]. However, human T. gondii infection may be associated with the consumption of raw or undercooked beef according to a previous epidemiological study [17]. Therefore, beef continues to highlight the crucial role cattle play in the transmission of this disease to humans.
Although the results of the present study were not significant, the prevalence of T. gondii infection in cattle tended to increase with their age. Similarly, other studies have demonstrated age-related increases in T. gondii seroprevalence in cattle [6,7,13]. This indicates that age is a risk factor for T. gondii infection because older individuals are exposed to the parasite for a longer duration than younger individuals [18]. With regard to sex, male cattle exhibited a significantly higher prevalence of T. gondii infection than female cattle in the current study, which is consistent with the findings of a previous study [14]. However, a higher prevalence in females was observed in another study [13]. In the present study, only brown cattle, raised for meat as beef cattle, were infected with T. gondii, although there was no significant correlation between infection and breed. Compared with dairy cattle, agricultural cattle, including brown cattle, inhabit a greater variety of habitats and thus may be more susceptible to contracting T. gondii infection [1]. In the present study, a significantly higher rate of T. gondii infection was observed in smaller farms than in larger farms, indicating that farm size is another risk factor. Similar findings from a different study revealed that the proportion of cattle from small farms with positive results was approximately 3 times higher than that of cattle from larger farms [13].
In a previous study, Toxoplasma strains were classified into 3 genotypes based on the virulence levels of the strains in outbred mice: type I (highly virulent strain), type II (less virulent strain), and type III (avirulent strain) [19]. Type II is the most prevalent genotype among humans [20]. In other studies, several types of T. gondii were identified in cattle, including types I (7.1%), II (70%), and III (17.2%) in Iran [14] and types I (1 sample) and II (4 samples) in Poland [13]. In the present study, 3 cattle tested positive for T. gondii, and using the B1 gene, we were able to identify the types I and/or III. Further phylogenetic analyses are required to differentiate between types I and III.
In conclusion, we detected a relatively low prevalence of T. gondii infection (0.7%) in cattle blood. To the best of our knowledge, this is the first molecular evidence of T. gondii in Korean cattle. Some aspects of the epidemiology of T. gondii infection in cattle remain unclear. Because cattle contain fewer tissue cysts than other meat-producing species, their role in the transmission of parasites to humans is questionable. However, the consumption of raw beef varies by region; the risk posed by T. gondii infection in cattle should not be underestimated. The risk of human infection can be mitigated by informing and educating customers regarding proper meat preparation and handling practices, with a focus on appropriate heat treatments for beef.